Chickpea Ascochyta: latest research on variability and implications for management
Author: Kevin Moore, Kristy Hobson, Nicole Dron, Steve Harden and Sean Bithell (DPI NSW), Prabhakaran Sambasivam (Uni of Melbourne), Rebecca Ford, Yasir Mehmood (Griffith Uni), Jenny Davidson (SARDI), Shimna Sudheesh and Sukhjiwan Kaur (Agribio DEDJTR). | Date: 01 Mar 2016
Take home message
- In 2015, Ascochyta blight occurred in a higher proportion of chickpea crops (60 of 243 crop inspections) than in 2014 (62 of 332 crop inspections). Most infected crops were PBA HatTrick which was also the most commonly grown variety.
- Work to determine if the Ascochyta pathogen is changing started in 2013, where a number of projects are working together to provide an integrated approach to chickpea Ascochyta blight to improve variety resistance and best management practices.
- Initial results show that the population varies in time for spore germination, germ tube length, ability to cause disease (pathogenicity), and time to develop fruiting bodies (latent period).
- Significant differences in the reaction of some varieties and advanced breeding lines to two aggressive isolates of the AB pathogen have been found
- It is essential that growers adhere to best management practices, such as sustainable rotations, to minimise selection pressure on the pathogen and maximise the longevity of variety resistance.
- While research into variability of the AB pathogen continues, it seems prudent to adopt a conservative approach to AB management
Ascochyta blight in 2015 chickpea crops
In 2015, 243 chickpea crop inspections were conducted as part of DAN00176. Ascochyta blight (AB) (Phoma rabiei formerly called Ascochyta rabiei) was detected in 60 crops. Inoculum had carried over from the 2014 season and wet conditions during Jun/Jul favoured infection and disease development. High chickpea prices tempted some growers to break best practice eg plant back to back chickpeas resulting in severe disease. Some growers reported more AB in PBA HatTrick than they ever saw in Jimbour but many of these crops had been inundated in Jun/Jul and we know that AB resistance of waterlogged chickpeas is compromised. Further the genetic purity of the variety could not be determined. Generally, however, good management and dry conditions through Aug – Oct kept AB under control and no major yield losses were reported.
Details of chickpea diseases and a review of the 2015 chickpea season are in another paper in these Proceedings (Chickpeas – what we learnt in 2015 and recommendations for 2016).
Latest research on variability in the Ascochyta pathogen
Is the pathogen changing? Yes, and as a population of living individuals (isolates), we should expect it to change.
Has the pathogen changed in response to selection pressure such as the widespread cultivation of varieties with improved resistance or other factors? We don’t yet know. To know if something has changed, you need to track it over a suitable time period. Detailed studies on molecular variability in the AB fungus commenced in 2008 and have shown that the overall population variation hasn’t changed much. However, pathogenicity studies that began in 2013 indicate that there are differences in pathogenicity among isolates and that highly pathogenic isolates are causing disease on PBA HatTrick. This paper provides key results from a range of research groups working on this combined project to better understand the chickpea AB population and its threat to the resistance sources through potential adaptation and selection.
Latent period
The incubation period is the time from infection to the appearance of symptoms. The latent period (LP) is the time from infection to the development of pycnidia (the small dark fruiting bodies that develop in the leaf and stem lesions), the LP is important because it determines how fast the disease can cycle in a crop. Determining these characteristics is thus another way of measuring variability in the pathogen population.
Three experiments were conducted in 2015. In each experiment, five isolates representing a sub-set of the pathogen population in Eastern Australia plus a 6th control isolate (obtained in 2014 from PBA HatTrick at Yallaroi, TR6415) were evaluated in a growth cabinet (20°C/15°C 12h day/12h night) on four chickpea genotypes. There were eight replicates (pots) for each of the 24 genotype by isolate combinations. At the 3 leaf stage plants were grouped by isolate and inoculated with a conidial suspension of 100,000 conidia/mL (sprayed to run-off). Plants were examined daily for symptoms and pycnidia. The mean LP was estimated by survival analysis with the status of a pot based on whether pycnidia had or had not developed. For each genotype-isolate, the data is the last day that pycnidia had not developed.
The four genotypes, their AB rating and abbreviation are: 1) ICC3996 (rated R, coded ICC), 2) GenesisTM 090 (rated R, coded GEN), 3) PBA HatTrick (rated MR, coded HAT), 4) Kyabra (rated S, coded KYB).
For each experiment, LP varied significantly between some isolates and genotypes (LP range 6-8 days). Furthermore, all isolates had the shortest LP on the most susceptible entry, KYB and the longest LP on the most resistant entry, ICC or the second most resistant entry, GEN (see example findings, Figure 1). Within an experiment, no single isolate had the shortest LPs on all genotypes, we interpret this as indicating there are no clear differences among isolates in the contribution of LP to isolate aggressiveness.
These experiments complement the pathogenicity work and confirm variability does exist in the pathogen population
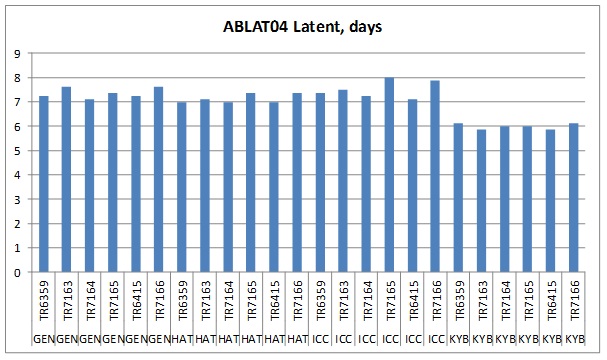
Figure1. Latent period results for experiment ABLAT04 grouped by genotype (ICC3996 (ICC), Genesis 090 (GEN), PBA HatTrick (HAT), Kyabra (KYB)) for inoculation with six isolates listed by isolate no, source and variety: TR6359 2014 North Star NSW, Flipper; TR7165 2014 Horsham VIC; Genesis425, TR7163 2014 Donald VIC; Slasher; TR6415 2014 Yallaroi NSW, HatTrick; TR7164 2014 Donald VIC, Slasher; TR7166 2014 Salter Springs SA, Monarch.
Histopathology experiments
A range of preliminary histopathology experiments have been completed, see Figure 2 for summary spore germination and germ tube length results. Key findings from a range of work in this area are that:
- Spore germination begins much faster on the susceptible Kyabra and on PBA HatTrick than on the resistant Genesis090
- Spore germination is consistently slower and lower on the resistance source ICC3996 than on any other chickpea genotype tested
- There is significant variation in germination time among different isolates and this correlates with their level of pathogenicity
- After germination, germ tube length prior to invasion is significantly shorter on ICC3996 than any other chickpea genotype tested
These differential fungal responses may be indicative of host recognition and defence strategies, which are being further investigated.
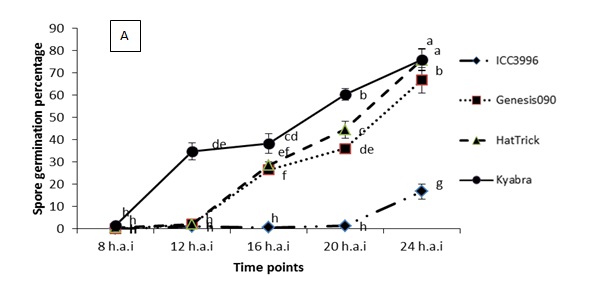
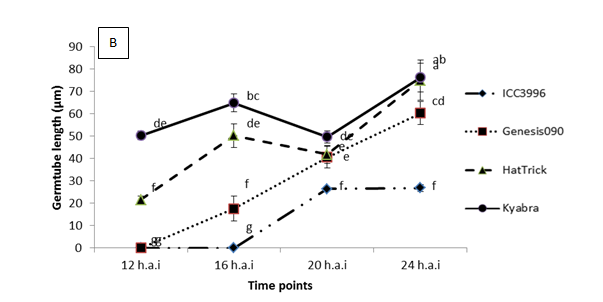
Figure 2. Significant differences were observed among the physiological traits of a highly pathogenic isolate FT13092-1 from Kingsford, SA when inoculated onto chickpea genotypes that are resistant (ICC3996 and Genesis090), moderately resistant (PBA HatTrick) or susceptible (Kyabra). Where A = the percentage of germinated spores and B = the germtube length over time after inoculation.
How is this information used by the PBA Chickpea program?In 2014 and 2015 two aggressive isolates identified by the pathogen variability project were screened on the national Stage 3 desi and kabuli entries in a controlled environment by SARDI. In 2015 the two isolates tested were collected in 2013; FT13092-1 from South Australia on Genesis 090 and TR5919 from northern NSW (Tooraweenah) on PBA HatTrick. Of the 154 entries tested, 62 breeding lines significantly differed in their resistance (% of main stem broken) to the two isolates (subset of lines presented in Table 1). The northern isolate was found to be more aggressive than the South Australian isolate. There was no significant difference in the response of PBA HatTrick to the two isolates, but PBA Boundary, CICA0912 and CICA1007 had significantly higher disease with TR5919. Conversely, the kabuli variety Genesis Kalkee had significantly lower disease with the TR5919 isolate compared to the SA isolate. The desi CICA1521 and kabuli CICA1156 had very low levels of disease from both isolates. The 2014 research examined two isolates collected in 2010 and a much smaller number of entries 8 (out of 137) had a significantly different response to the two isolates.
To complement this information, molecular markers have been screened across the 154 entries. A total of 5 flanking molecular markers (3 SNPs and 2 SSRs) for AB resistance (resistance sources S95362 (kabuli) and ICC3996 (desi)) were identified within “DAV00098 - Molecular markers for the pulse breeding programs” led by DEDJTR, Victoria. These markers have been validated across a diverse set of chickpea lines as part of DAV00126 program. By combining the phenotypic and genotypic information, the breeding program will gain a greater understanding of the genetic resistance in each breeding line. The wider implementation of AB molecular markers across the PBA Chickpea program has identified breeding material which may contain alternative resistance genes. Research into alternative genetic resistance genes is continuing in DAV00126. The use of alternative resistance genes in the breeding program will be essential to ensure new chickpea varieties have adequate levels of AB resistance.
Table 1. Ascochyta blight ratings, response of varieties and breeding lines (% main stems broken, lsd 29.2) to two Phoma rabiei isolates in a controlled environment and presence/absence (+/-) of molecular marker and source of resistance.
|
AB |
% of main stems broken |
|||
|
Name |
Field rating |
Isolate FT13092-1 |
Isolate |
Marker genotype |
|
Kyabra |
S |
100 |
100 |
- |
|
PBA HatTrick |
MR |
0 |
20 |
+, desi |
|
PBA Boundary |
MR |
35 |
75 |
+, desi |
|
Genesis 836 |
MS |
8 |
28 |
Not conclusive |
|
CICA0912 |
R* |
0 |
42 |
+, desi |
|
CICA1007 |
MR* |
0 |
50 |
+, desi |
|
CICA1521 |
R* |
0 |
8 |
+, desi |
|
Almaz |
MS |
8 |
8 |
-, suggests other genes |
|
Genesis 090 |
R |
0 |
8 |
+, kabuli |
|
Genesis 425 |
R |
8 |
17 |
+, kabuli |
|
Genesis Kalkee |
MS |
50 |
20 |
--, suggests other genes |
|
PBA Monarch |
MS |
3 |
42 |
+, kabuli plus others |
|
CICA1156 |
R* |
0 |
0 |
+, kabuli |
*Advanced breeding lines, putative AB rating
While research into variability of the AB pathogen continues, it seems prudent to adopt a conservative approach to AB management
Further information
GRDC Factsheet Chickpea disease management 2013
- Ascochyta blight in chickpea
- Northern chickpea disease management guide
- Chickpea IDM strategies
- Botrytis grey mould in chickpeas
- Chickpea phytophthora root rot
- Sclerotinia in chickpeas
- Managing viruses
and in the NSW DPI 2016 Winter Crop Variety Sowing Guide
Acknowledgements
This research would not be possible without the considerable and ongoing support from growers and the GRDC for which we are most grateful. Thanks to Paul Nash and Gail Chiplin for technical support. Thanks also to agronomists for help with the crop inspections and submitting specimens.
Contact details
Kevin Moore
NSW DPI
Ph: 02 6763 1133
Mb: 0488 251 866
Fx: 02 6763 1100
Email: kevin.moore@dpi.nsw.gov.au
Kristy Hobson
Ph: 02 6763 1174
Mb: 0400 955 476
Fx: 02 6763 1100
Email: kristy.hobson@dpi.nsw.gov.au
® Registered trademark
Varieties displaying this symbol beside them are protected under the Plant Breeders Rights Act 1994.
GRDC Project Code: DAN00176, UM00052, DAN00151, DAV00126, DAN00151, DAV00098,
Was this page helpful?
YOUR FEEDBACK
