The incidence of fungicide resistance in spot form net blotch (SFNB) and its implications
Author: Fran Lopez-Ruiz, Wesley Mair, Geoff Thomas, Kith Jayasena and Andrea Hills | Date: 25 Feb 2019
Key Messages
- Fungicide resistance to Group 3 (DMI) fungicides in spot form of net blotch is spreading in the southern region of WA.
- Overuse of fungicides with the same mode of action will speed up fungicide resistance.
- We can limit the development of fungicide resistance by using the lowest effective dose, appropriate fungicide group rotations and employing IDM practices including crop rotation, stubble management and selection of more resistant cultivars (if available).
- Fast (and cheap) monitoring of pathogen populations for fungicide resistance is central for the sustainable chemical management of diseases.
Aims
Broadacre cropping is an important Australian industry, contributing over $21 billion of the gross value of agricultural production in 2016-17 (ABS 2018). Foliar fungal diseases are an important limiting factor to broadacre yield and quality production worldwide. Nowadays, the control of fungal diseases in broadacre crops is very reliant on the use of broad spectrum single-site fungicides. However, the continuous application of fungicides has led to the development of resistance in many fungal pathogens, reducing the management alternatives available for growers and threatening profitability.
A small number of systemic actives from a single mode of action (MOA) group – the DMIs or Group 3 fungicides - dominate the market. Although other important systemic and contact fungicides from different MOA groups have been introduced in recent years, the overreliance on chemical disease control strategies, has led to the quick development of fungicide resistance in Australian fields. So far, five cases of fungicide resistance and three cases of reduced sensitivity (resistance that does not reach the level of field failure) have been identified in Australia since 2012 (table 1).
Barley spot form net blotch (SFNB) is caused by Pyrenophora teres f. maculata and is the most significant barley necrotrophic disease in Australia (Murray and Brennan, 2010). Along with cultural practices, the main control measures are the application of effective fungicides and the use of cultivars with genetic resistance. However, due to the lack of highly resistant cultivars, SFNB is mainly controlled using fungicides (Sierotzki et al., 2007).
The efficacy of some Group 3 fungicides has been impacted by the development of resistance in SFNB in WA populations. This paper provides an overview of the current knowledge on SFNB fungicide resistance and the spread of the resistant populations in WA.
Table 1. Fungicide resistant diseases found in Australian grains. Current at 10th January 2019.
Disease | Fungicide Group | Compounds affected | Region | Industry implications |
|---|---|---|---|---|
Barley powdery mildew | 3 (DMI) | Tebuconazole, propiconazole, flutriafol | WA, Qld, NSW, Vic, Tas | Field resistance to some Group 3 fungicides |
Wheat powdery mildew | 3 (DMI) | None | NSW, Vic, Tas | This is a gateway mutation. It does not reduce the efficacy of the fungicide but is the first step towards resistance evolving |
11 (QoI) | All group 11 | Vic, Tas | Field resistance to all Group 11 fungicides | |
Barley net-form of net blotch | 3 | Tebuconazole, propiconazole, prothioconazole, epoxiconazole | WA | Reduced sensitivity that does not cause field failure |
Barley spot-form of net blotch | 3 (DMI) | Tebuconazole, propiconazole, prothioconazole, epoxiconazole | WA | Field resistance to some Group 3 fungicides |
Canola blackleg | 2 (MAP-Kinase) | Iprodione | WA | Not registered for this disease but used against diseases that share the same host |
3 | Fluquinconazole | WA, NSW, Vic, SA | Field implication unclear | |
Wheat septoria leaf blotch | 3 | Tebuconazole, flutriafol, propiconazole, cyproconazole, triadimenol | NSW, Vic, SA, Tas | Reduced sensitivity that does not cause complete field failure |
Chocolate spot | 1 (MBC) | Carbendazim | SA | Field resistance to carbendazim |
Ascochyta blight | 1 | Carbendazim | SA | Field resistance to carbendazim |
Source: Fran Lopez-Ruiz, Angela van de Wouw, Andrew Milgate, Nick Poole and Richard Oliver. GRDC project codes CUR00016, CUR00023, UM00051 and DAN00177.
Method
Fungal samples and in vitro fungicide resistance analysis
Four hundred and ninety-five samples of SFNB diseased barley were collected by a combination of especially designed bait trials (86 samples), field trips (127 samples) and a network of collaborators (282 samples) from farms and trials during the 2016, 2017 and 2018 growing seasons. Bait trials located in Inverleigh, Vic, Esperance, WA, Northam, WA and Mt Barker, WA were sown with the SFNB susceptible varieties SY Rattler, Fathom, Spartacus, Stirling and Planet and were designed to work as a fungicide resistance catch system. Plots of 2m × 4m were sprayed with either 1× or 2× the maximum registered dose of fungicides from the Groups 3 (DMI), 11 (QoI) and 7 (SDHI), at growth stages GS31 and GS39. Treatments were replicated three times. Leaf samples from bait trials were collected seven days following the second spray application.
Samples were processed in the laboratory and pure fungal isolates established following the isolation procedure described by Mair et al. (2016). A set of fungicide discriminatory concentrations was established based on previous analysis of the sensitivity baselines of 23 SFNB isolates collected between 1995 and 2013 to different fungicides (tebuconazole = 10µg/mL; epoxiconazole = 5µg/mL; boscalid = 10µg/mL; azoxystrobin = 5µg/mL). Isolates able to grow above those concentrations were considered to be resistant in vitro. Isolates able to grow above discriminatory concentrations were subjected to further in vitro fungicide sensitivity analysis, by growing the cultures at different concentration ranges of the fungicides, in order to determine the concentration of each fungicide that inhibits the growth of each isolate by 50 per cent, also known as effective concentration 50 (EC50). Resistance factors (RF) were determined as fold number difference between EC50 values from resistant isolates and the average of the 23 sensitive isolates. High RF, EC50 values, or growth of the isolates at discriminatory doses does not necessarily imply field failure – further studies are needed.
Molecular analysis of fungicide resistance
Isolates able to grow above discriminatory concentrations were subjected to digital Polymerase Chain Reaction (dPCR) analysis for the detection of the known fungicide resistance mutation F489L. This mutation affects the binding of the Group 3 fungicide to the target site only. In addition to the isolate analysis, DNA extracted from infected barley samples were screened for the presence of the mutation using this same methodology. The assay uses specific molecular probes to detect the presence of the mutation in diseased plant material, without the need to isolate pure strains of the pathogen. This not only greatly reduces turnaround time but also allows accurate quantitation of the frequency of the mutation in the sample, and the detection of the mutation at very low frequencies (>0.2%).
SFNB isolates that grew above Group 3 discriminatory doses were also subjected to further molecular characterisation. The Group 3 fungicide target site gene was sequenced in resistant isolates in order to determine if other mutations were associated with the resistance levels detected. Where mutations were found that may affect the over-production of the fungicide target, the production level of target gene of the resistant isolates were measured and compared to that of the wild types.
A second dPCR detection methodology was developed for the detection of certain small DNA fragment insert mutations associated with moderately-resistant SFNB. Leaf samples from field trials and from collaborators collected in the 2018 growing season were screened using this method.
Results
Group 3 fungicide resistance in SFNB
P. teres f. maculata with reduced levels of sensitivity to several DMI fungicides were detected in samples from Western Australian from 2016 onwards (Table 2). In vitro testing sorted strains into sensitive (S), moderately resistant (MR), and highly resistant (HR) groups based on 50% effective fungicide concentrations (EC50) to the Group 3 compounds propiconazole, prothioconazole, tebuconazole, prochloraz, difenoconazole and metconazole. The MR and HR groups showed a similar level of reduced sensitivity to epoxiconazole (Table 3).
Table 2. Number of SFNB-infected barley field samples tested in each year, showing proportions in which Group 3 sensitive, moderately resistant or highly resistant strains were detected. aFrom 2018 season onwards, field samples were routinely screened for the presence of resistance mutations using molecular detection methods. Frequencies are shown in brackets.
Population | 2016 | 2017 | 2018a | Total |
|---|---|---|---|---|
S | 48 (92.3) | 213 (98.2) | 170 (75.2) | 431 (87.1) |
MR | 4 (7.7) | 1 (0.5) | 43 (19) | 48 (9.7) |
HR | 0 (0) | 3 (1.4) | 13 (5.8) | 16 (3.2) |
Total | 52 | 217 | 226 | 495 |
Table 3. Mean effective concentration 50 (EC50) in µg/ml of five Group 3 moderately resistant isolates, fifteen highly resistant isolates, and a reference population of 23 sensitive SFNB isolates collected between 1995 and 2013. Resistance factors (fold number difference between EC50 values from resistant isolates and average of sensitive isolates) are given in brackets. Cultures were grown at different concentration ranges of the fungicides tebuconazole, epoxiconazole, prothioconazole and propiconazole. aMean of 23 sensitive SFNB isolates; bmean of a subset of 3 sensitive SFNB isolates.
Population | Tebuconazole | Epoxiconazole | Prothioconazole | Propiconazole |
|---|---|---|---|---|
Sensitive (1995 – 2013) | 0.31a | 0.17a | 0.07a | 0.16b |
Moderately Resistant (MR) | 2.56 (8.6) | 1.72 (10.6) | 0.49 (6.8) | 0.46 (2.9) |
Highly Resistant (HR) | 16.69 (55.9) | 1.45 (8.9) | 1.67 (22.9) | 6.8 (43.2) |
Analysis of thetarget gene for the group 3 fungicide, called Cyp51A, revealed the presence in MR and HR isolates of two different mutations that were not observed in sensitive isolates. In MR isolates, the mutation was a small fragment of DNA that was inserted in the fungicide target gene. This small fragment of new DNA was found at three different positions and its effect was the over-production of the fungicide target. The presence of more fungicide target requires an increase in the amount of fungicide necessary to kill the MR isolates.
The small DNA fragment was also found in HR isolates but at a different position, together with another mutation, F489L. This latter mutation has been previously observed in the closely related pathogen P. teres f. teres, the causative agent of net form of net blotch (NFNB), where it has been correlated with reduced sensitivity to a range of DMI fungicides (Mair et al. 2016).
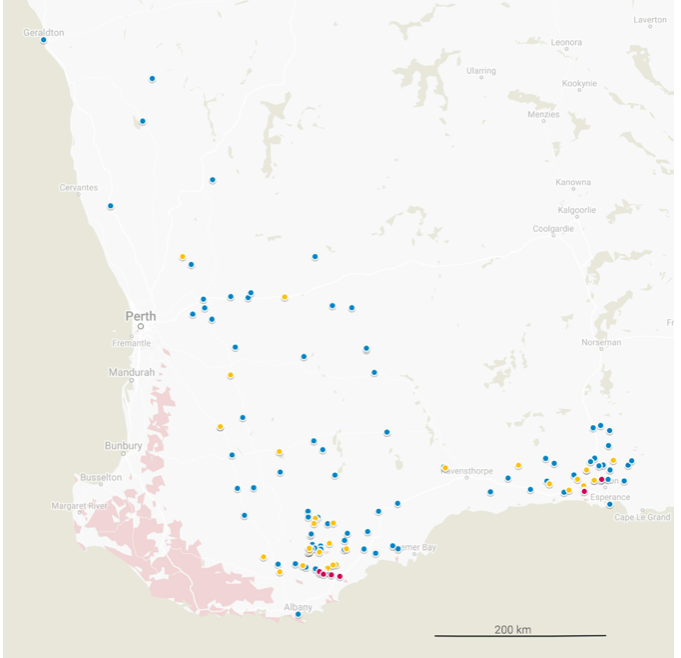
Moderately resistant strains were isolated from samples collected in field trips in the Esperance region (Gibson, Coomalbidgup and Cascade) from 2016 onwards, and highly resistant isolates were derived from samples collected by collaborators in the Great Southern (South Stirling and Wellstead) and Esperance (Dalyup) regions from 2017 onwards (Figure 1). A dPCR assay for certain DNA inserts associated with moderate resistance in SFNB was able to detect the presence of the mutation in samples from several additional sites across WA in the 2018 growing season (Figure 1).
Conclusion
With the development of resistance to Group 3 fungicides in SFNB, growers need to be cautious about its control and implement adequate integrated disease management strategies to minimise the ongoing selection of SFNB resistant populations. Being a stubble borne disease, rotating crops or managing stubble are paramount for reducing disease carryover and selecting disease resistant varieties will reduce disease severity. However, these measures will not be very effective unless care is given to the choice of fungicide. Any spray program heavily dependent upon Group 3 fungicides will increase the selection of the resistant populations. The introduction of seed dressing, in-furrow and foliar products containing fungicide mixtures from different chemical groups (Groups 3, 7 and 11) in combination with the removal of tebuconazole from the control programs in those areas where resistance is found, will provide the best opportunity to limit the spread of the resistance in SFNB and its emergence in other barley growing regions of Australia.
Management
In addition to using cultivars with good disease resistance levels, clean seeds, stubble management strategies to reduce disease load, crop rotation, grazing and maintaining good farm hygiene, the following chemical management strategies are recommended:
* Only spray if necessary – limit applications.
* Choose mixtures with different modes of action (if available).
* Never apply the same Group 3 fungicide twice in a row.
* Avoid applying the same mode of action fungicides from the Groups 7 (SDHI) and 11 (Qol) twice.
* Incorporate the use of seed dressing (Group 7), in-furrow (Group 11) and foliar products containing fungicide mixtures from different chemical groups (such as 3 (DMI), 7 (SDHI) and 11 (Qol)) - in combination with limited use of propiconazole and no stand-alone tebuconazole use.
* Avoid using tebuconazole as a stand-alone product in barley for any disease to avoid indirect fungicide resistance selection.
* Use other DMI-based mixtures (such as propiconazole, prothioconazole or epoxiconazole) only once, followed by mixtures containing other actives (preferably from groups 7 or 11).
* If resistance is present, or suspected, avoid or minimise use of that mode of action - this will only further select for resistance.
* Do not exceed label rates.
Acknowledgments
The research undertaken as part of this project is made possible by the significant contributions of growers through both trial cooperation and the support of the GRDC, the author would like to thank them for their continued support.
The authors would like to acknowledge as well the contribution of Department of Primary Industries and Regional Development (DPIRD, WA), The Foundation for Arable Research in Australia (FAR) and the grains industry.
Special thanks go to growers and grower groups submitting samples to the Fungicide Resistance Group (FRG) and contributing to our research.
GRDC Project Number: CUR00016, CUR00023
References
ABS (2018), Australian Bureau of Statistics
Murray, G.M.,and Brennan, J.P. (2010). Estimating disease losses to the Australian barley industry. Aust. Plant Pathol. 39,85–96.doi:10.1071/AP09064
Sierotzki, H., Frey, R., Wullschleger, J., Palermo, S., Karlin, S., Godwin, J., et al. (2007). Cytochrome b gene sequence and structure of Pyrenophora teres and P. tritici-repentis and implications for QoI resistance. Pest Manag. Sci. 63, 225–233.doi:10.1002/ps.1330
Mair, W.J., Deng, W., Mullins, J.G.L., West, S., Wang, P., Besharat, N., Ellwood, S.R., Oliver, R.P. and Lopez-Ruiz, F.J. (2016). Demethylase Inhibitor Fungicide Resistance in Pyrenophora teres f. sp. teres Associated with Target Site Modification and Inducible Overexpression of Cyp51. Front. Microbiol. 7:1279. doi: 10.3389/fmicb.2016.01279
Was this page helpful?
YOUR FEEDBACK
