Mapping Ascochyta rabiei aggressiveness and understanding the pathogen adaptation to disease management strategies
Mapping Ascochyta rabiei aggressiveness and understanding the pathogen adaptation to disease management strategies
Author: Ido Bar (Griffith University) | Date: 25 Feb 2022
Take home message
Ascochyta rabiei isolates are getting more aggressive in recent years and are able to overcome some of the `resistant` chickpea varieties used by the industry.
Unique clusters of highly aggressive isolates have been identified that potentially pose a major and ongoing risk to the industry.
The risk from local micro-evolution of highly aggressive isolates becomes even greater when combined with cross-region gene flow that is likely driven by human activities.
To be able to offer the most accurate and up-to-date information on the spread of Ascochyta blight and provide early warning of the emergence of highly aggressive A. rabiei isolates, industry action is needed. Action needed is in the form of; adherence to the use of clean seed, machinery hygiene practices and maximum frequencies for chickpeas in the rotation to be established and adhered to.
Engage with pathologists when Ascochyta blight symptoms are seen to assist in sample collection and pathogen monitoring.
Introduction
Research on the Australian Ascochyta rabiei population over the past eight years has established a comprehensive isolate database, provided insights into the population structure and identified trends in adaptation and possible modes of evolution across all chickpea growing regions. A subset of the collected isolates in each year are phenotyped against a differential host set of chickpea genotypes (ICC3996, Genesis™ 090, PBA HatTrick, PBA Seamer and Kyabra) to determine their potential aggressiveness (classified as Pathogenicity Group 0-5, from the least aggressive to the most aggressive). The phenotyped isolates are further characterised molecularly and genotyped to determine their mating type, genetic relatedness and the population structure.
Ascochyta rabiei in Australia appears to be clonal (which means the offspring are identical to their parent), with detection to date of a single mating type (MAT1-2). Despite this form of reproduction, which is suggested to limit new genetic diversity, it seems that the population contains sufficient existing diversity to undergo spontaneous mutation events to adapt to and overcome host resistance sources. This is evident by the detection of severe disease symptoms on the formerly released ‘resistant’ hosts, such as Genesis™ 090, PBA Seamer and ICC3996 (the resistance source for many commercial varieties, see Figure 1).
Since 2015 there has been an increase in the frequency of collected isolates that can cause severe disease on ‘resistant’ cultivars. In particular, the proportion of isolates highly aggressive on PBA HatTrick rose from 18% in 2013 to 51% in 2020 (Figure 1). Similarly, a higher frequency of isolates able to cause severe disease on Genesis™ 090 was observed (up from 11% in 2016 to 36% in 2020). This is particularly concerning since PBA HatTrick and Genesis 090 are broadly sown cultivars in northern and southern chickpea regions, respectively. Of most concern for northern growers is the growing number of highly aggressive isolates on PBA Seamer (up from 4% in 2016 to 48% in 2020, Figure 1) (Sambasivam et al., 2020).
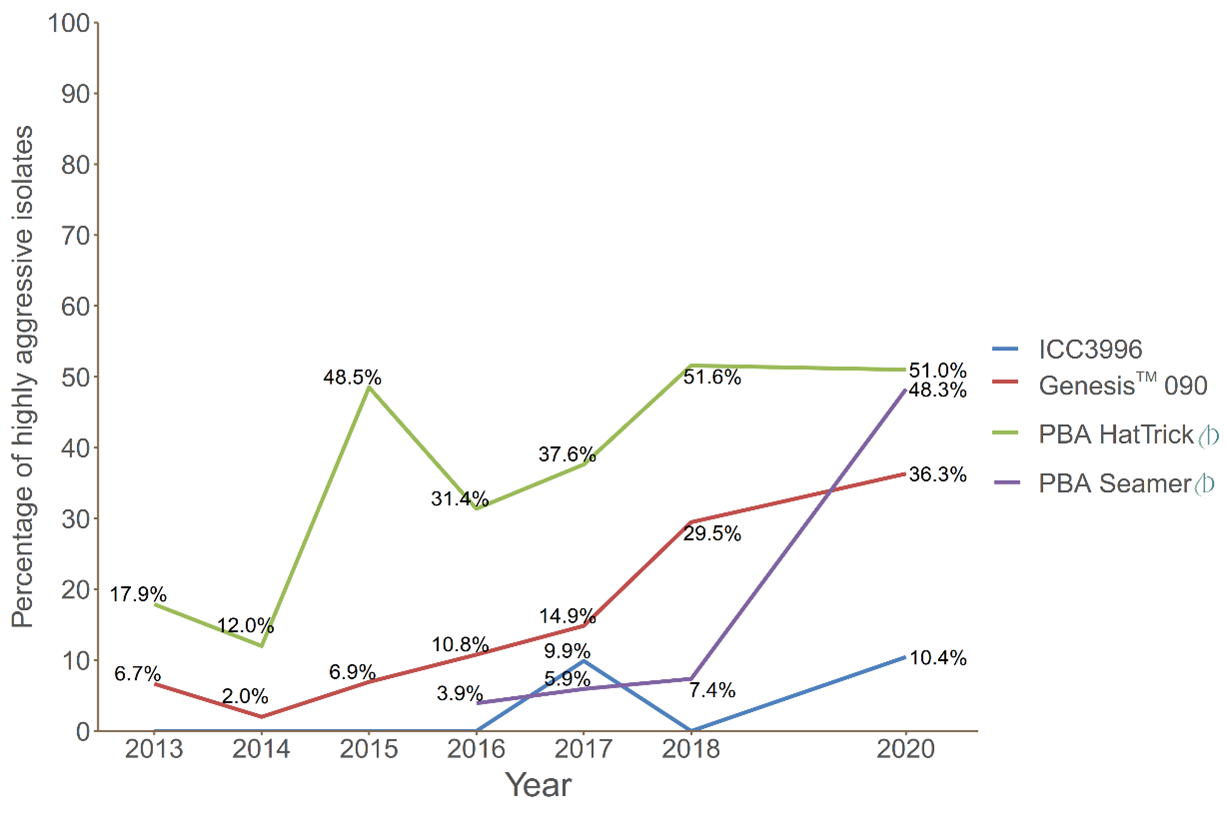
Figure 1. Percentages of Ascochyta rabiei isolates screened in controlled environment bioassays able to cause severe disease on the four chickpea hosts assessed (95-200 isolates screened
per year)
A high-resolution genetic analysis of 193 isolates collected from all growing regions in 2020 discovered unique genetic clusters of highly aggressive isolates within specific locations that potentially pose a major and ongoing risk to the industry. Isolates in these clusters are 10-13 times more likely to be highly aggressive than other isolates. Based on the genetic relatedness of the isolates we identified two likely sources that contribute to the emergence of new isolates: either local micro-evolution of isolates that can lead to adaptation of highly aggressive isolates or cross-region gene flow likely driven by human activities. Both mechanisms pose a major and ongoing risk to the industry. Identifying the source of new genetic material in an event of an outbreak is crucial to be able to manage the risk and control the disease effectively (Bar et al., 2021).
To support informed disease management strategies and identify trends and patterns affecting the emergence of highly aggressive isolates, an online dashboard was developed and deployed as a live web application to enable interactive interrogation of A. rabiei isolates collected within the current (and previous) GRDC investments. The dashboard provides a spatio-temporal overview of 437 isolates that were collected in 2020-2021, along with specific details of their collection metadata. It also details the phenotypic assessment and summary plots of 806 isolates that were phenotyped between 2013-2020. The dashboard was developed with an interactive and user-friendly interface and is available at http://bit.ly/asco-dashboard
These findings and further advances in the understanding of the pathogen populations have significant implications for development of accurate diagnostics tools and informed disease management strategies at a regional and national scale to assist Australian scientists, breeders, farmers and government agencies in managing and reducing the chickpea Ascochyta blight risk for sustainable production of chickpea.
References
Bar I, Sambasivam PT, Davidson J, Farfan-Caceres LM, Lee RC, Hobson K, Moore K and Ford F (2021) Current population structure and pathogenicity patterns of Ascochyta rabiei in Australia. Microbial Genomics 7, 000627
Sambasivam P, Mehmood Y, Bar I, Davidson J, Hobson K, Moore K and Ford R (2020) Evidence of recent increased pathogenicity within the Australian Ascochyta rabiei population. bioRxiv 2020.06.28.175653 doi:10.1101/2020.06.28.175653.
Acknowledgements
The research undertaken as part of this project is made possible by the significant contributions of growers through both trial cooperation and the support of the GRDC, the author would like to thank them for their continued support.
We would also like to thank the various state government pulse pathologists for their assistance in the sample collection, as well as to other members of the investment (CSIRO, SARDI, CCDM, ICARDA, DJPR) for their collaboration and sharing of knowledge.
Contact details
Dr. Ido Bar
Griffith University
170 Kessels Road, Nathan QLD 4111
Ph: 0435 718 770
Email: i.bar@griffith.edu.au
Melody Christie
Griffith University
170 Kessels Road, Nathan QLD 4111
Ph: 0435 718 770
Email: m.christie@griffith.edu.au
Varieties displaying this symbol beside them are protected under the Plant Breeders Rights Act 1994.
GRDC Project Code: GRI2007-001RTX,
