Inoculum detection capability affects disease risk prediction for Phytophthora root rot of chickpeas
Author: Sean Bithell (NSW DPI), Kristy Hobson (NSW DPI), Steve Harden (NSW DPI), Willy Martin (DAFQ), Steven Simpfendorfer (NSW DPI), Alan McKay (SARDI) and Kevin Moore (NSW DPI) | Date: 06 Mar 2018
Call to action/take home messages
- For soil samples where Phytophthora medicaginis (Pm) concentrations are low (< 1500 Pm DNA sequences/g soil), PREDICTA® B was variable both in detection success (the number of positive samples) and estimating Pm concentration.
- Pm DNA was not detected at the lowest inoculum concentration treatment (32 oospores/plant) in a Pm inoculated field trial; Phytophthora root rot (PRR) disease did develop in these plots, but only at low levels.
- The ability of soil Pm DNA concentrations to predict yield losses depends on soil water conditions. Under dry spring conditions, little PRR developed in an inoculated field trial despite high inoculum treatments. However, irrigated treatments in the same trial led to increasing disease incidence with increasing inoculum concentrations and associated yield losses.
- Detecting any level of Pm in a paddock using PREDICTA® B makes that paddock a high risk of developing PRR if conditions become conducive. However, not detecting Pm does not mean the PRR risk is low.
- The Pm DNA PREDICTA® B test is useful as an in-crop diagnostic tool for growers and agronomists to confirm PRR disease; the best diagnostic results will be from plants collected as they are dying when the pathogen is active and inoculum concentrations are high.
Phytophthora medicaginis detection and quantification in soil
Phytophthora medicaginis (Pm), the cause of chickpea Phytophthora root rot (PRR) is endemic and widespread in the northern grains region. Under conducive conditions, PRR can cause 100% loss. The pathogen survives from season to season on chickpea volunteers, lucerne, native medics, sulla and as resting structures (oospores) in roots and soil. Pm inoculum (infected root tissue and oospores) is difficult to detect and quantify in paddocks when a susceptible host such as chickpeas is not present (Dale and Irwin, 1990).
A PREDICTA® B soil DNA test has been developed by the South Australian Research and Development Institute (SARDI) to quantify the amount of Pm DNA in soil samples and so provide a measure of the amount of Pm inoculum in paddocks. We report on the 2017 season of studies to assess the capability of this test to:
- Detect Pm in soil samples
- Predict the risk of PRR disease development and potential yield losses in chickpea
Pm DNA detection in soil samples
We evaluated the ability of the Pm DNA test to detect Pm in soil naturally infested with Pm across a dilution series. Previous work on the Pm PREDICTA® B tool has shown that soil samples collected from within chickpea crops with PRR problems, would consistently give positive detection results (Bithell et al 2015 & 2016 Update papers). But detection in break crops gave low Pm detection success even for specific sites (‘hotspots’) in paddocks where ongoing PRR problems had recently been documented (Bithell et al 2017 Update paper). Pm detection success across an oospore-mycelium concentration series indicated that variable detection success may occur in samples where concentrations are low (≤0.25 oospores/g soil) (Bithell et al 2017 Update paper).
This raises a key question for break crop sampling: was the low success of detecting Pm in soil samples due to the pathogen population being below a consistent level of detection?
Soil was collected from under naturally PRR infected chickpea cv Sonali plants from a field trial (CPRRNATTW16) at Tamworth, NSW. The soil was sieved (4 mm aperture), thoroughly mixed, then diluted with sand to give seven concentrations (Table 1); with four replicates. The samples were sent to SARDI for Pm DNA analysis.
The number of positive detections declined as the dilution with sand increased. In terms of the determination of Pm concentration, with the exception of the 12.5% dilution, the average number of Pm DNA sequences of all four samples also declined as the dilution with sand increased. One aspect of the results for the average number of Pm DNA sequences calculated only from positive samples, there was no consistent decline in sequence numbers between the 25% soil and 3% soil treatments. These results indicate that in naturally infested soils where Pm concentrations are relatively low (<1500 Pm DNA sequences/g soil), variability in both detection success and Pm concentration is expected.
Table 1. Soil dilution series results, percentage and mass (g) of naturally Pm infested soil, number of Pm DNA sequences/g soil and number of positive DNA detections.
% soil in sample | g of soil in sample | Average no. Pm DNA sequences/g soil – all samples | No. of positive samples | Average no. Pm DNA sequences/g soil – positive samples |
|---|---|---|---|---|
100 | 300 | 2372 | 4/4 | 2372 |
50 | 150 | 1785 | 4/4 | 1785 |
25 | 75 | 1013 | 3/4 | 1350 |
12.5 | 37.5 | 258 | 2/4 | 515 |
6.3 | 18.8 | 451 | 1/4 | 1804 |
3.1 | 9.4 | 277 | 1/4 | 1109 |
0 | 0 | 0 | 0/4 | 0 |
Disease and yield loss prediction trial: ability to predict disease severity and yield loss
It would be useful to growers if the Pm DNA test could predict the likely extent of PRR disease development and potential yield losses within paddocks. For example, would paddocks with nil, low and high Pm inoculum levels respectively have nil, low and high PRR disease development and yield losses? In addition, could low inoculum levels at sowing produce similar disease to high inoculum levels, if conditions during the growing season were conducive i.e. wet soil (as provided by irrigation in this experiment)?
This was the aim of a 2017 field trial conducted at the DAF Q Hermitage Research Facility, Warwick using the MR rated chickpea variety, Yorker. On 28 June 2017, a range of Pm inoculum levels (Table 2) was established by applying different rates of oospores (a mixture of 10 isolates) in-furrow at sowing. Pooled soil samples (32 x 150 mm depth cores) from the middle two rows of each plot were collected at sowing and analysed for soil Pm concentrations. Establishment counts were conducted on 31 August and PRR disease incidence (number of chlorotic and dead plants) assessments were made throughout season, with final grain yield measured at harvest (11 December). The trial was split for dryland (D; no irrigation) and irrigation (I); the latter applied in the first week of August and third week of October with dripper tape.
Soil Pm DNA levels established at sowing differed significantly when measured using PREDICTA® B and increased with the targeted inoculum concentration treatments, although no DNA was detected in the lowest inoculum treatment (32 oospores/plant; Table 2). Uniform plant populations (average 20.4 plants/m of row) were established across the trial with no significant effect of inoculum or watering treatments (data not shown). The growing season was drier than usual (137 mm from 29 June to 30 Nov with only 7 mm total for August and Sept compared to 284 mm average from 1994 to 2017 for this period). However, by 31 August, PRR symptoms (wilting and chlorosis) were observed in most treatments.
At the end of September, significant trends were evident for increasing disease incidence with increasing inoculum concentrations (Table 2). Disease incidence was also higher in the irrigated treatments compared with dryland at the two highest inoculum treatments. Although the incidence of diseased plants generally increased with increasing inoculum concentration, there was no correlation with grain yield in the dryland treatments (Table 2, Figure 1). In contrast, under irrigation yield significantly declined across groups of inoculum treatments, with 0 & 32 oospores > 164 & 360 oospores > 668 & 1683 oospores > 3270 oospores (Table 2, Figure 1).
Table 2. Inoculum level, sowing soil Pm DNA concentration, PRR incidence and yield in 2017 trial (D = dryland, I = irrigated)(soil Pm concentration P < 0.001; lsd soil Pm concentration = 982.5; 23 Sept. number of PRR diseased plants <0.05; lsd no diseased plants 19.7; Grain yield <0.001, lsd Yield = 598.6).
Inoculum level (oospores/plant) | Pm DNA concentration (no. Pm sequences/g soil) | Disease incidence | Grain yield (kg/ha) | ||
|---|---|---|---|---|---|
D & I | D & I | D | I | D | I |
0 | 0 | 0 | 0 | 1907 | 4357 |
32 | 0 | 5 | 10 | 2562 | 4044 |
164 | 285 | 18 | 35 | 2128 | 3317 |
360 | 363 | 24 | 40 | 2028 | 3281 |
668 | 863 | 55 | 71 | 1485 | 2582 |
1683 | 1081 | 35 | 66 | 2302 | 2936 |
3271 | 2858 | 55 | 108 | 2136 | 1402 |
Results from this experiment highlight the importance of soil moisture conditions in the relationship between soil Pm DNA concentrations at sowing and yield loss. Low rainfall in August and September (7 mm – i.e. 58 mm below average) limited the ability for PRR to develop to damaging levels under dryland conditions despite high inoculum concentrations in some dryland treatments. Another aspect affecting the predictive ability of soil Pm concentrations measured at sowing is the relatively wide range of DNA results obtained across a single targeted oospore concentration treatment (Figure 1). This highlights the inherent sensitivity and sampling issues using the Pm PREDICTA® B te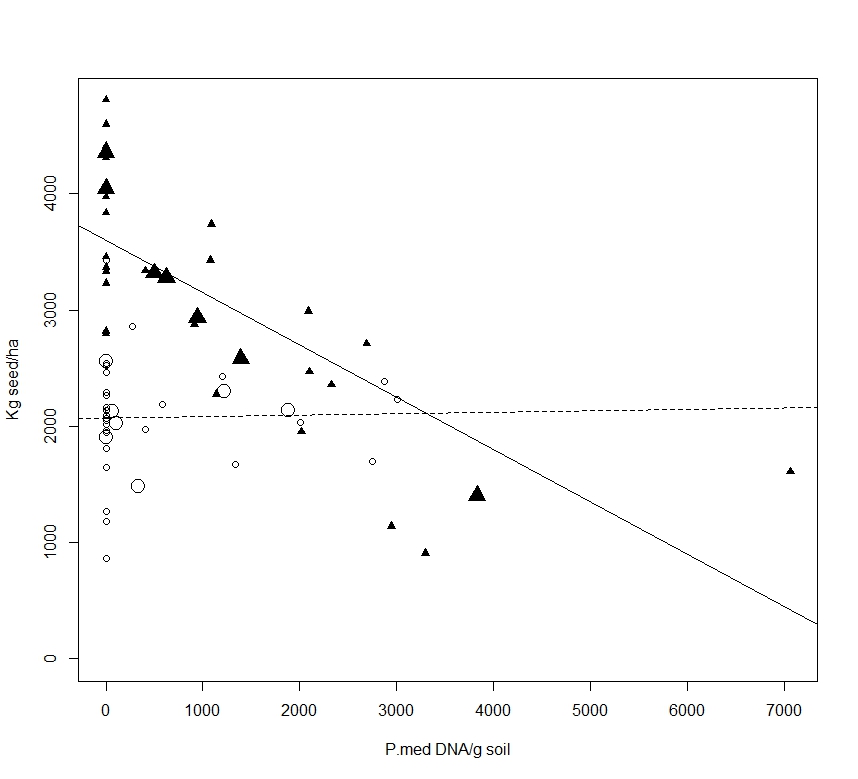
Figure 1. Post sowing soil Pm DNA concentrations for irrigated (filled triangle symbols) and dryland (open circle symbols) treatments verses Yorker grain yields (kg/ha), average oospore inoculum treatments are large symbols, small symbols are actual plot values. Lines represent linear regression for irrigated (solid line, grain yield = 3599-0.449(DNA), r2 0.52, P = 0.903) and dryland (broken line, grain yield = 2072-0.0013(DNA), r2 0.0005, P < 0.001) treatments.
Implications for growers
The low success of the Pm PREDICTA® B test to consistently detect inoculum in paddocks where Pm concentrations are low, makes the test unreliable at low inoculum levels as a predictor of disease and yield loss. This is because under conducive soil water conditions, the disease may develop from low or undetectable levels of inoculum and still cause significant damage.
However, the Pm DNA test is useful as an in-crop diagnostic tool for growers and agronomists to confirm the presence of PRR disease. For example, in 2016 some chickpea paddocks in NW NSW were saturated causing some areas of the paddocks to die. Pm DNA analysis of soil samples from some of these areas has allowed agronomists and growers to identify if waterlogging and/or PRR were the primary cause(s) of the losses.
Confirmation of PRR in chickpea crops through isolation of the pathogen from diseased tissue, can be unsuccessful if the symptoms are advanced, or the plants have died due to colonisation of dead tissue by other soil microbes. Hence if PREDICTA® B is to be used as a diagnostic tool in-crop, soil and plants samples will still need to be collected as plants are dying when the pathogen is active and inoculum concentrations are high.
In summary, in terms of our key questions:
Identify if a low Pm detection success in soil samples may be due to the pathogen population being below a consistent level of detection.
We found that variability in both detection success and Pm concentration may be expected in soils with low (< 1500 Pm DNA sequences/g soil) inoculum concentrations. Samples from chickpea break crops have either nil Pm or Pm concentrations below 1000 Pm DNA sequences/g soil (Bithell et al 2016 Update paper), so it is expected that PRR risk prediction based on low Pm DNA results in this range will be unreliable.
Would paddocks with nil, low and high Pm inoculum levels respectively have nil, low and high PRR disease development and yield losses?
Our field trial results show that Pm inoculum concentrations will not predict yield loss for the MR variety Yorker in a low rainfall season, especially with very limited spring rainfall. However, where soil moisture conditions are higher throughout a season, then inoculum concentrations can be used to predict yield loss with adequate (r2 = 0.52) but not high accuracy.
Would conducive conditions (wet soil provided by irrigation) allow low inoculum levels at sowing to produce similar disease development to high inoculum levels?
We found no evidence that wet soil provided by irrigation provided the ability for low inoculum levels at sowing to produce similar disease development to high inoculum levels for the MR variety Yorker. However, irrigation with dripper tape in this trial may not provide conditions as conducive as natural rain events and greater PRR development may occur in seasons with average or high rainfall.
Acknowledgements
This research was a co-investment between NSW DPI and GRDC under project DAS00137. The ‘ability to detect Pm in paddock samples’ research was partly funded by project DAN00172. We thank growers for the significant contributions through both trial cooperation, paddocks access and the support of the GRDC. Assistance provided by Gail Chiplin, Paul Nash and Amy Trebilco (NSW DPI) is greatly appreciated.
References
Bithell, S., Moore, K., Hobson, K., Harden, S., and A. McKay (2015) A new DNA tool to determine risk of chickpea Phytophthora – in paddocks. Grains Research Update, Goondiwindi, QLD, 3-4 March 2015. Grains Research and Development Corporation. 167-171.
Bithell, S., Moore, K., Hobson, K., Harden, S., Martin, W., and A. McKay (2016) A new DNA tool to determine risk of chickpea Phytophthora root rot. Goondiwindi, QLD, 1-2 March 2016. Grains Research and Development Corporation. 92-96
Bithell, S., Harden, S., Hobson, K., Martin, W., and K. Moore (2017). Disease risk prediction for Phytophthora root rot of chickpeas: inoculum detection problems. Grains Research Update Dubbo, 28 Feb-1 March. Pages 121-128.
Dale, M. L. and J. A. G. Irwin (1990). Estimation of inoculum potentials of Phytophthora megasperma f.sp. medicaginis in chickpea fields and the development of a glasshouse resistance assay. Australian Journal of Experimental Agriculture 30: 109-114.
Contact details
Sean Bithell
NSW Department Primary Industries
Ph: 0429 201 863
Fx: 02 6763 1100
sean.bithell@dpi.nsw.gov.au
Kevin Moore
NSW Department Primary Industries
Ph: 0488 251 866
Fx: 02 6763 1100
kevin.moore@dpi.nsw.gov.au
® Registered trademark
Was this page helpful?
YOUR FEEDBACK
