Using deoxyribonucleic acid (DNA) markers to improve wheat breeding
Author: Adam Norman (PhD candidate, University of Adelaide; Australian Grain Technologies) | Date: 12 Feb 2019
Take home messages
- Until recently, DNA markers have been ineffective when breeding for complex traits such as grain yield.
- Genomic selection is a strategy which uses thousands of DNA markers to predict the performance of new breeding lines.
- This research has identified effective and efficient strategies for implementing genomic selection, and increasing the rate of genetic gain achieved.
Background
DNA markers have been employed in plant breeding for decades. Despite early optimism around their ability to achieve genetic gain for important traits such as yield, they have so far only proved valuable for simple traits such as rust resistance, and been of more use as a research tool for understanding genetic control of various traits. Genomic selection is a technology that has garnered significant research attention in recent years following its successful implementation in the dairy cattle breeding industry. Genomic selection utilises high density DNA marker profiles to develop prediction calibrations for complex traits such as yield, which allow us to predict the performance of new breeding lines before they are assessed in the field. Such an ability would enable plant breeders to deliver greater genetic gain to growers through increased speed and accuracy in the breeding programme. Genomic selection can be applied at various stages of a breeding programme, as detailed in Figure 1.
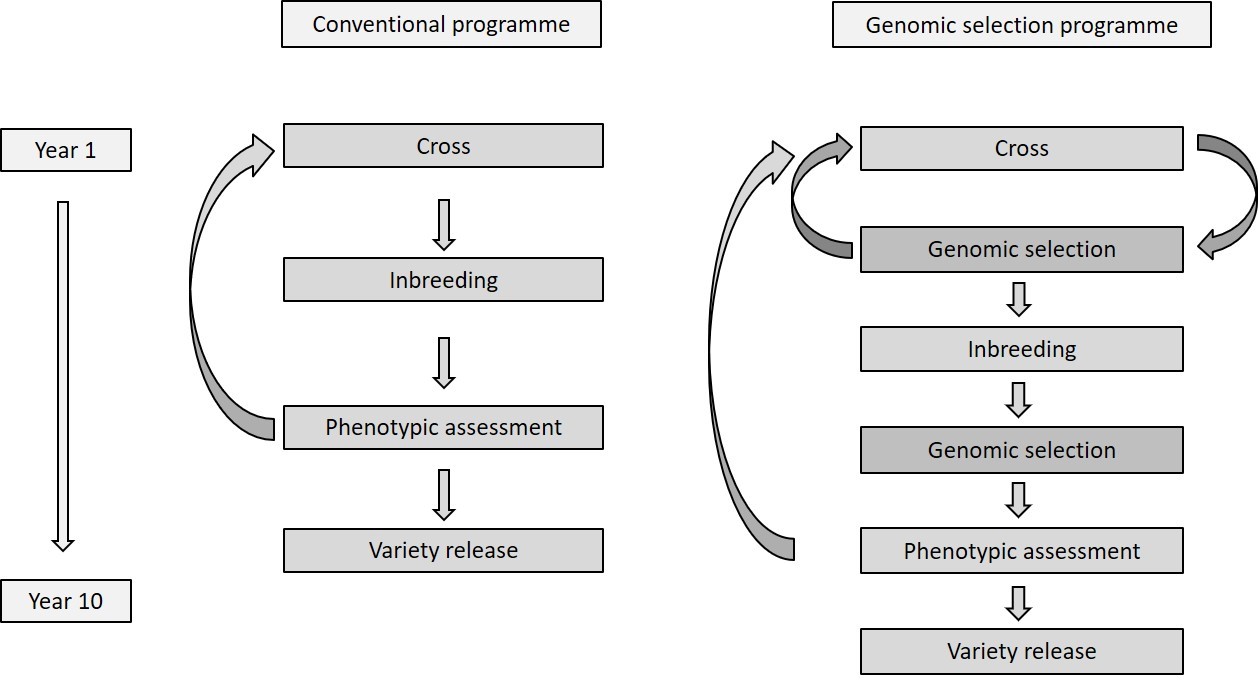
Figure 1. Comparison of conventional and genomic selection breeding programmes, highlighting the points in the programme where genomic selection can be applied.
For genomic selection to be successful, prediction calibrations must capture the full genetic variation of the breeding programme and also account for the range of target environments. It is therefore important that we understand the requirements of the data used to build the prediction calibrations and know the limitations of how broadly they can be applied. Much of the previous research on this topic has employed relatively small training set sizes of less than 1000 individuals. However, if genomic selection is to be applied in commercial breeding programmes, training set sizes could exceed 10,000 individuals. Genomic selection must therefore be studied in larger datasets if its efficacy is to be proven.
Project outputs
A large panel of 10,500 wheat breeding lines was used to develop genomic prediction calibrations and assess their accuracy. The panel was genotyped with 18,101 DNA markers and phenotyped for 15 traits in a field experiment at Roseworthy in the 2014 growing season. The relationship between training set size and prediction accuracy was investigated where it was found that prediction accuracies continue to benefit from increased training set sizes up to the largest tested (8300 individuals), showing that very large datasets are required to effectively implement a genomic selection breeding strategy. The effect of population structure was also investigated and showed that a smaller training set can achieve sufficiently high prediction accuracies if the genetic variation of the germplasm of interest is strongly represented in the training set.
Acknowledgements
The research undertaken as part of this graduate research project is made possible by the significant contributions of growers through their support of the GRDC, the author would like to thank them for their continued support. We also thank the Australian Research Council for their support through funding, and Australian Grain Technologies for their significant in-kind contribution.
Contact details
Adam Norman
Australian Grain Technologies
0400 656 012
adam.norman@agtbreeding.com.au
@adamnormanAGT
GRDC Project Code: UA00152,
Was this page helpful?
YOUR FEEDBACK
