New tool to predict the likelihood of an inoculation response to Group E and F rhizobia
Author: Ross Ballard, Stephen Barnett, Daniele Giblot-Ducray, Herdina, Kelly Hill, Elizabeth Farquharson and Alan Mckay (SARDI and University of Adelaide) | Date: 09 Feb 2021
Take home messages
- A new DNA soil testing service, PREDICTA® rNod has been developed to measure Group E and F rhizobia numbers in soil to assist growers to identify the need to inoculate field pea, faba bean, lentil and vetch crops.
- PREDICTA rNod is available to South Australia (SA) and Victoria (Vic) growers via PREDICTA® B accredited agronomists in 2021 and will be launched to growers nationally in 2022.
- The DNA test will be a valuable research tool to investigate how soil chemistry and management practices affect the survival of Group E and F rhizobia in soil, nodulation and pulse performance.
- DNA tests for chickpea and lupin rhizobia are also currently under development.
Background
The number of rhizobia present in soil is a key determinant of legume nodulation, growth and nitrogen fixation and where rhizobia numbers are low, they must be delivered in inoculants applied to the seed or soil.
Rhizobia levels in soil are affected by the frequency of host pulse crops, soil type, soil pH and high temperatures. The lack of a reliable way to estimate rhizobia populations has resulted in many growers applying inoculants as an ‘insurance policy’, with some of this inoculation likely to be ineffectual. On the other hand, some growers don’t inoculate where rhizobia levels are too low which will compromise the nodulation of the pulse crop. Recent expansion of the pulse industry is seeing crops grown in new and marginal environments that are responsive to rhizobial inoculation.
A DNA test that can accurately and rapidly estimate the number of Rhizobium leguminosarum bv. viciae in the soil has been developed for growers and researchers. This is the species of rhizobia provided in the commercial inoculant Groups E (strain SU303) and F (strain WSM-1455). The test will help growers identify paddocks where field pea, faba and broad bean, lentil and vetch crops will need to be inoculated before sowing or not.
When the test indicates soil rhizobia levels are adequate, growers will be able to consider applying fungicides or trace elements to the seed and/or dry sowing, knowing these practices pose a negligible risk to legume nodulation.
The test will also enhance research capacity to understand how inoculation and agronomic practices (e.g., liming or rotation) influence rhizobia number in the soil and affect the performance of pulse crops.
This paper describes the development and evaluation of the DNA test that measures the number of Rhizobium leguminosarum bv. viciae, in soil.
Methods
The new rhizobia DNA test is based on a qPCR assay (TaqMan MGB), specific for Rhizobium leguminosarum bv. viciae (hereafter referred to as E and F rhizobia).
Specificity of the test was investigated using DNA extracted from 83 cultures of rhizobia, comprising 42 strains of E and F rhizobia and 41 strains of non-target closely and distantly related rhizobia (Table 1).
Sensitivity of the DNA test and correlation with viable rhizobia numbers per gram of soil were determined by calibrating the DNA test against the Most Probable Number (MPN) plant nodulation bioassay (Vincent 1970), using vetch cv. Timok as the trap plant. This study used 41 field soil samples collected from cereal stubbles between December 2019 and February 2020.
Nitrogen (N) fixation capacity of the soil rhizobial communities was also determined in a greenhouse experiment. Shoot dry weight of field pea (cv. Kaspa) and faba bean (cv. Samira) reliant on the soil rhizobia for growth was compared with the shoot dry weight of plants inoculated with commercial inoculant strains SU-303 (Group E for pea) or WSM-1455 (Group F for bean).
Spatial variation of E and F rhizobia across paddocks, and on and off row was measured in three grower paddocks using the DNA test, to determine suitable paddock sampling strategies.
Results and discussion
Specificity of the DNA test
A DNA test has been designed to detect only E and F rhizobia that nodulate field pea, faba bean, lentil and vetch (Table 1). The test does not detect strains of the closely related clover rhizobia (Rhizobium leguminosarum bv. trifolii). This is important because both biovars often coexist in many paddocks. Additionally, the test does not detect more distantly related rhizobia.
Table 1. Specificity results (detected, not detected) for the DNA test targeting E and F rhizobia, includes 14 rhizobia species and 83 strains.
Legume (rhizobia Group) | Strains tested | Detected | Not detected |
|---|---|---|---|
Pea, vetch, bean, lentil (Group E & F) | 42 | 42 | 0 |
Clovers - close relative (Group B & C) | 20 | 0 | 20 |
Medic and lucerne (Group AM & AL) | 8 | 0 | 8 |
Ten other rhizobia species | 13 | 0 | 13 |
Sensitivity and correlation to viable rhizobia in soil
The DNA and MPN plant bioassay methods were significantly correlated across 41 soil samples.
The MPN test was more sensitive at lower levels of rhizobia (<200 rhizobia/g soil), with several instances of detection by the plant bioassay, but not the DNA test (Figure 1).
The DNA test was able to reliably detect around 1,000 cells/g soil and was more precise than the MPN test when rhizobia DNA levels exceeded this level. Overall, there was a high correlation (adjusted R2=0.86) between the log transformed measures of the MPN and DNA tests.
As few as 100 rhizobia/g soil are sufficient to nodulate pulse crops in the field, similar to the number applied with peat inoculant on seed. The testing service will use conservative thresholds; soils with >1,000 to 5,000 rhizobia/g (log10 >3 to 3.7) will be assigned a low likelihood of response to rhizobia inoculation, and >5,000 rhizobia/g soil to indicate a negligible likelihood of inoculation response.
Based on previous surveys of E and F rhizobia in soils (Drew et al. 2012), it was expected that around 30% of soils will be classified as having a low or negligible likelihood of response to inoculation with E and F rhizobia.
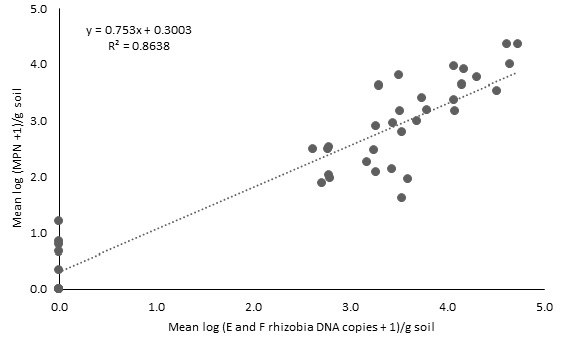
Figure 1. Relationship between E and F rhizobia DNA copies/g soil measured by DNA test and rhizobia number/g soil measured by Most Probable Number (MPN) plant nodulation bioassay, for 41 soils collected between December 2019 and February 2020.
For the soils tested, several unexpected results were noted. These included instances of high E and F rhizobia numbers (2,971/g soil) despite no known history of a pulse host crop and conversely, low rhizobia numbers (111/g soil) despite inoculated faba bean having been grown in 2015. These variations underline the value of the new test in helping better target the inoculation of pulse crops.
Nitrogen fixation capacity of soil rhizobia
Ten of the field soils in Figure 1 were estimated to contain more than 1,000 E and F rhizobia/g soil (after conversion from raw DNA data), and therefore, were predicted to have a low likelihood of response to rhizobia inoculation. The N-fixation capacity of the rhizobial communities in these soils with field pea and faba bean is shown in Table 2.
With field pea, N–fixation capacity ranged for 64 to 99% relative to inoculant strain SU-303, and with faba bean from 39 to 107% relative to WSM-1455. With the exception of Soil 10, the N-fixation capacity of the soil communities should be high enough to supply sufficient fixed N for field grown plants, because plant growth rate and demand for N is lower in the field compared to plants grown in the greenhouse. Even though the rhizobia in Soil 10 were less effective, the site would be unlikely to respond to inoculation due to competition from the soil rhizobia community, so the DNA test prediction of a low likelihood of inoculation response remains reasonable.
Table 2. Nitrogen fixation capacity of E and F rhizobia (Rhizobium leguminosarum bv. viciae)communities predicted by the DNA test to exceed 1000/g soil. N-fixation capacity calculated as % of shoot dry weight for field pea inoculated with SU-303 and faba bean inoculated with WSM-1455.
Soil | DNA copy number #/g soil | Predicted MPN E/F rhizobia/g soil | N-fixation capacity Pea, % SU-303 | N-fixation capacity Bean, % WSM-1455 |
|---|---|---|---|---|
Soil 1 | 49,201 | 6,792 | 74 | 104 |
Soil 2 | 47,284 | 6,592 | 92 | 107 |
Soil 3 | 32,331 | 4,951 | 69 | 78 |
Soil 4 | 20,728 | 3,542 | 72 | 85 |
Soil 5 | 16,410 | 2,971 | 99 | 107 |
Soil 6 | 13,380 | 2,547 | 70 | 84 |
Soil 7 | 12,710 | 2,451 | 70 | 89 |
Soil 8 | 11,493 | 2,272 | 97 | 76 |
Soil 9 | 4,740 | 1,166 | 94 | 73 |
Soil 10 | 4,187 | 1,062 | 64 | 39 |
Soil sampling
Several aspects of soil sample collection have been examined to develop suitable sampling protocols.
Spatial variation across paddocks
Where there are obvious differences in soil type or management zones within a paddock, each zone should be sampled separately for testing. This requirement is illustrated in the following examples:
- Samples from areas in a paddock that differed in pHCa (5.0 or 7.1) also varied in predicted E and F rhizobia number, 288 and 2,971/g soil, respectively.
- Samples collected from hills and flats in a paddock, varied in predicted E and F rhizobia number, 375 and 1,062/g soil, respectively.
In both examples, E and F rhizobia numbers in the different zones would be assigned to different categories of inoculation requirement in the report generated for growers.
Where paddocks were ‘uniform’ in soil type and management, mean E and F rhizobia numbers were similar, regardless of whether the paddock was sampled as a whole (single test sample of 500g, 45 cores), as a split paddock (two 500g test samples) or from nine different sectors (Figure 2).
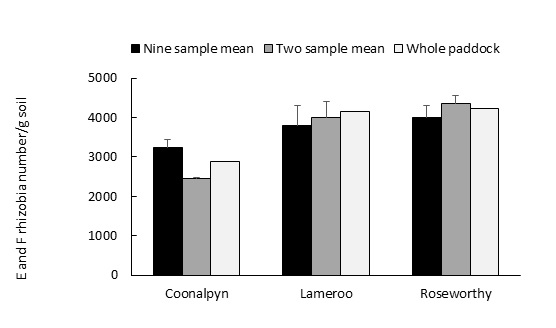
Figure 2. Effect of paddock sampling frequency (nine, two or one sample locations) at three field sites on mean predicted E and F rhizobia number/g soil. Bars above columns indicate standard error.
The effect of sampling within or between cereal stubble rows on the number of E and F rhizobia was also investigated. Levels were not different between locations (P = 0.218) in three different paddocks (Table 3). Hence, samples for the E and F rhizobia DNA test can be taken from either position.
Table 3. Effect of sample position, within or between rows of cereal stubble, on the predicted number of E and F rhizobia/g soil at three field sites. Each value is the mean of nine 500 g paddock samples.
Location | Predicted E and F rhizobia number/g soil (0-10 cm) | |
|---|---|---|
Within stubble rows | Between stubble rows | |
Coonalpyn, SA | 3,914 | 3,238 |
Lameroo, SA | 4,391 | 3,802 |
Roseworthy, SA | 4,832 | 3,991 |
Mean of all sites | 4,028 | 3,677 |
Time of sampling
Results to date have been based on dry soil samples collected in February preceding the pulse crop, by which time rhizobia populations have declined to levels approximating numbers persisting at the break of season.
Investigations are currently underway to determine if earlier sampling times (Oct-Nov) in the year before the pulse crop can provide a reliable estimate of rhizobial number persisting through to the next season. This timing will align with the timing of inoculant orders.
Recommended PREDICTA rNod soil sampling protocol
For ‘uniform’ production zones, it is recommended that a composite 500g soil sample made up of three cores (10 mm wide x 100mm depth) is collected at each of 15 locations within a production zone or soil type (total 45 cores/sample). Unlike disease testing, there is no need to target the rows of the last crop or to add stubble.
Research applications
The rhizobia DNA test for E and F rhizobia will improve the efficiency of research programs and provide agronomists with a tool to understand the impact of management practices, such as liming, on soil health. The test has already been used successfully to:
- Select trial sites free of rhizobia group E and F for inoculation experiments.
- Compare the colonisation of soils by different inoculant strains.
- Quantify nodulation on legume roots to compare management practices.
As commercial and research sample results become available, it will be possible to generate regional summaries of rhizobia status, on a scale that has not previously been possible.
Conclusion
A new DNA test has been developed to measure E and F rhizobia numbers in soil prior to sowing field pea, faba and broad bean, lentil and vetch.
The E and F rhizobia test will form the basis of a new service, PREDICTA rNod, which will be available to SA and Vic growers from February 2021, via PREDICTA B accredited agronomists.
The test will indicate that inoculation responses are unlikely when E and F rhizobia numbers exceed 1,000 cells/g soil. Soil pH and texture results will also be reported to assist with interpretation.
Soil samples can be collected from the start of February, when rhizobia numbers should approach levels persisting at the break of season. Use PREDICTA rNod kits to submit a composite 500g soil sample made up of three cores (10 x 100mm) collected at each of 15 locations within a production zone (total 45 cores/sample).
More soils are being added to the calibration data set to support the release of the test nationally.
Further work is also being undertaken to investigate earlier sampling for growers who want to know inoculant requirements at least six months before seeding a pulse crop.
Tests for chickpea and lupin rhizobia are under development and are expected to be released in 2022.
Acknowledgements
This research was initiated by the GRDC Bilateral Project DAS00137 and completed by the Bilateral Project UOA1802-019BLX. Calibration of the test has been undertaken by the GRDC funded Nitrogen Fixation Program UOA1805-017RTX (9176500). This is made possible by the significant contributions of growers through both trial cooperation and the support of the GRDC, the authors would like to thank them for their continued support.
The authors would also like to thank the following agronomists and growers for their support and provision of soil samples; Andy Bates, Michael Hind, Simon Mock, Jeff Braun, Mick Faulkner, Andrew Heinrich, Sam Trengove, Stuart Sherriff, John Matheson, Peter Maynard and Neville Kernick.
Useful resources
Inoculating Legumes: A Practical Guide:
PREDICTA B Agronomist Broadacre Soilborne Disease Manual V10.4:
PREDICTA rNod kits available from PREDICTA B accredited agronomists and Russell.burns@sa.gov.au
References
Drew EA, Denton MD, Sadras VO, Ballard RA (2012) Agronomic and environmental drivers of population size and symbiotic performance of Rhizobium leguminosarum bv. viciae in Mediterranean-type environments. Crop and Pasture Science. 63: 467-77.
Vincent JM (1970) ‘A manual for the practical study of root nodule bacteria.’ (Blackwell Scientific Publications: Oxford)
Contact details
Ross Ballard
SARDI Soil Biology and Diagnostics
GPO Box 397, Adelaide SA, 5001
+61 8 8429 2217
ross.ballard@sa.gov.au
Varieties displaying this symbol beside them are protected under the Plant Breeders Rights Act 1994
GRDC Project Code: UOA1805-017RTX (9176500),
Was this page helpful?
YOUR FEEDBACK
